Accurate predictions of conformational ensembles of disordered proteins
STARLING takes flight
Excited to announce the newest member of the flock - STARLING (conSTruction of intrinsicAlly disoRdered proteins ensembles efficientLy vIa multi-dimeNsional Generative models) (Novak* et al., 2025)
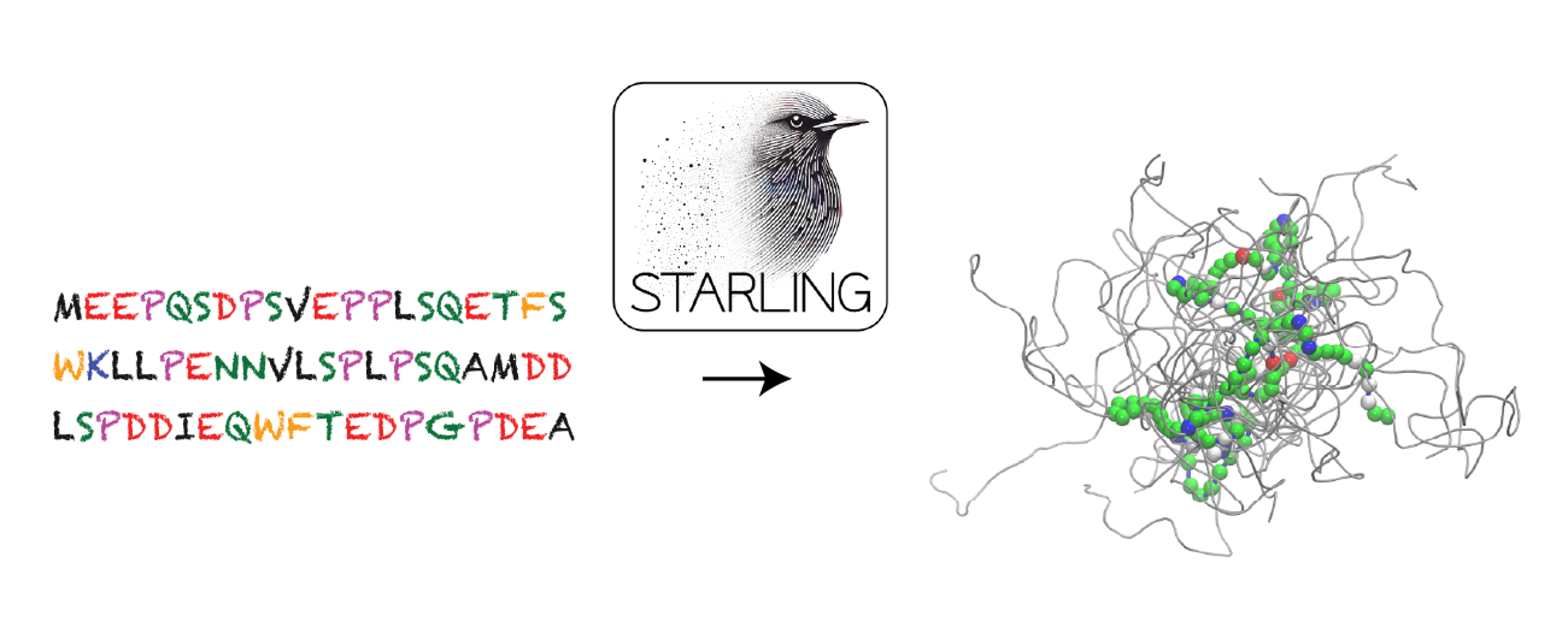
STARLING is a generative model for the accurate prediction of coarse-grained disordered protein conformation ensembles.
STARLING is a collaborative project spearheaded by Borna Novak and myself which builds upon the lab’s foundational work of IDR conformational ensemble property prediction directly from sequence (Lotthammer* et al., 2024).
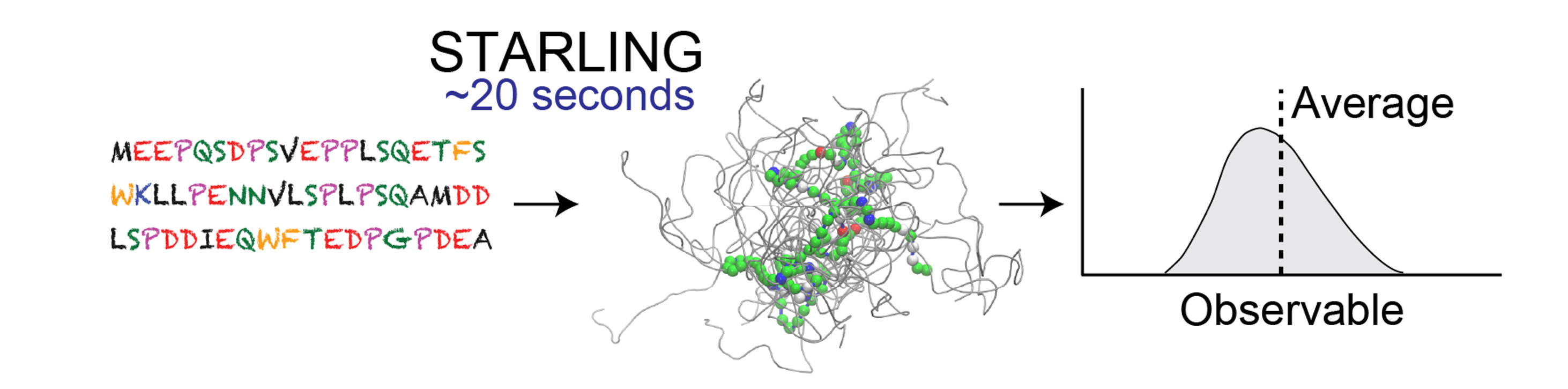
While previous deep learning approaches have focused on predicting average values for some subset of observables (e.g. end-to-end distance), they are limited by which observables have predictive models.
STARLING presents a generalization of this recent work by enabling the generation of IDR ensemble from which any observable and its distribution can be computed.
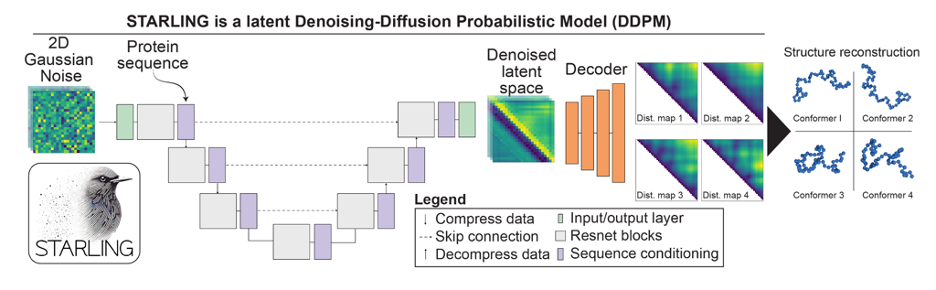
STARLING is a latent denoising diffusion model inspired by recent progress in text-to-image generative models.
We formulate IDR ensemble construction as a process of generating instantaneous distance maps in a sequence-conditioned manner, where each map represents a structure based on pairwise inter-residue distances.
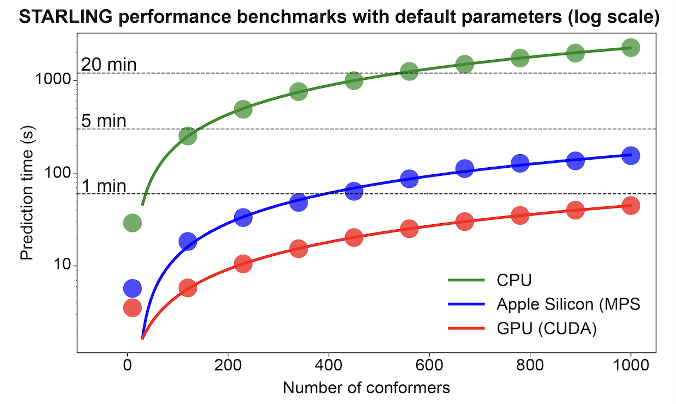
STARLING produces high-quality predictions at a blazingly fast rate on GPUs and Apple Silicon and is still performant on CPUs.
We benchmark STARLING against decades of elegant biophysical research of disordered proteins, including smFRET, SAXS, and NMR experiments, and find that STARLING displays remarkable agreement.
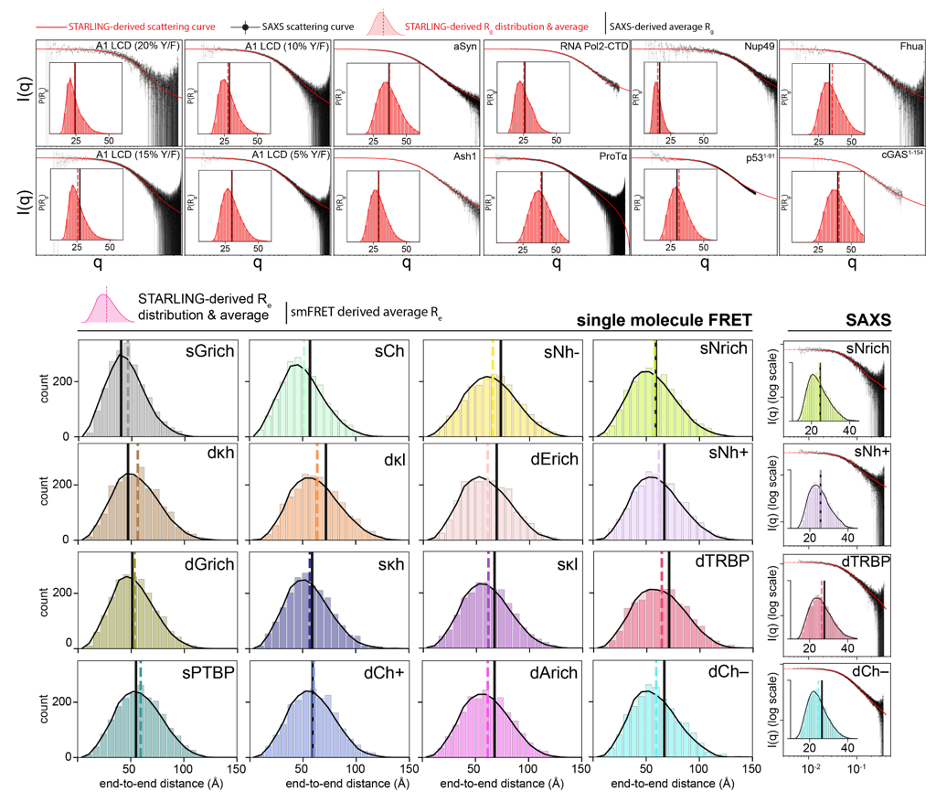
STARLING dramatically lowers the barrier to the computational interrogation of IDR function through the lens of emergent biophysical properties in addition to traditional bioinformatic approaches.
STARLING can be used to develop hypotheses as to how an IDR’s sequence may determine its conformational ensemble and/or how it may influence interactions with other IDRs.
We also show how one can integrate STARLING with protein design tools to build de novo disordered protein sequences with target ensemble properties.
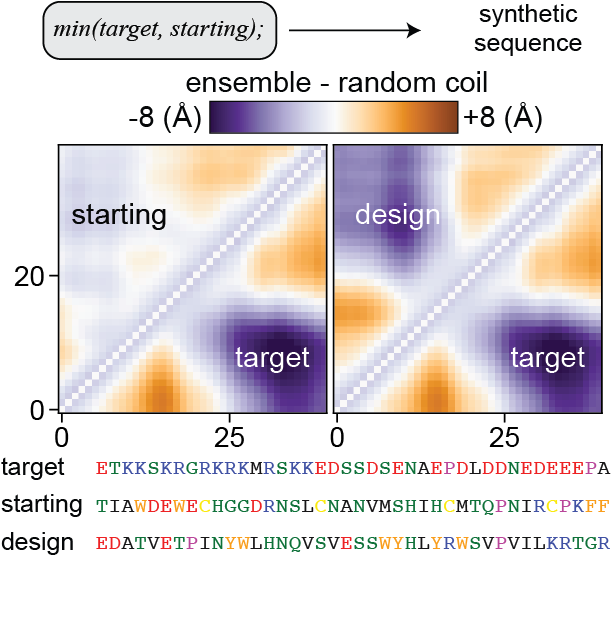
Importantly, STARLING is an open-source tool targeting ease of use and widespread availability. STARLING is available to install and run locally or online through a simple interface via Google Colab.
Installation
You should really go on over to the github for this information, but… since I’m here I wanted to give a little demo.
I recommend creating a fresh conda environment for STARLING (although in principle there’s nothing special about the STARLING environment)
conda create -n starling python=3.11 -y
conda activate starling
You can then install STARLING from GitHub directly using pip:
pip install idptools-starling
Or you can clone and install the bleeding-edge version from GitHub
git clone git@github.com:idptools/starling.git
cd starling
pip install .
To check STARLING has installed correctly run
starling --help
Quickstart
The easiest way to use STARLING is the starling command-line tool.
starling <amino acid sequence> -c <number of confomers> --outname my_cool_idr
This will generate an output file call my_cool_idr.starling. To convert this to a PDB trajectory run
starling2pdb my_cool_idr.starling
Or to convert to an xtc/pdb combo run:
starling2xtc my_cool_idr.starling
Python library
STARLING can generate Ensemble objects which enable deep investigation into ensemble properties using the generate function.
generate function documentation
The generate function is the main entry point for generating distance maps using the STARLING model. This function accepts various input types, generates conformations using DDPM, and optionally returns the 3D structures. You can customize several parameters for batch size, device, number of steps, and more.
To get started, first import the function:
from starling import generate
The generate function is flexible and can take in sequences in multiple formats. Here are a few examples:
# Example 1: Provide a single sequence as a string
sequence = 'MKVIFLAVLGLGIVVTTVLY'
# E is an Ensemble() object
E = generate(sequence, return_single_ensemble=True)
# Example 2: Provide a list of sequences
sequences = ['MKVIFLAVLGLGIVVTTVLY', 'MKVIFLAVLGLGIVVTTVLY']
# returns a dictionary of the Ensemble() objects
E_dict = generate(sequences)
# Example 3: Provide a dictionary of sequences
# returns a dictionary of the Ensemble() objects
sequences = {'seq1': 'MKVIFLAVLGLGIVVTTVLY', 'seq2': 'MKVIFLAVLGLGIVVTTVLY'}
E_dict = generate(sequences)
References
2025
-
 Accurate predictions of conformational ensembles of disordered proteins with STARLINGbioRxiv, Feb 2025
Accurate predictions of conformational ensembles of disordered proteins with STARLINGbioRxiv, Feb 2025
2024
-
 Direct prediction of intrinsically disordered protein conformational properties from sequenceNature Methods, Mar 2024
Direct prediction of intrinsically disordered protein conformational properties from sequenceNature Methods, Mar 2024